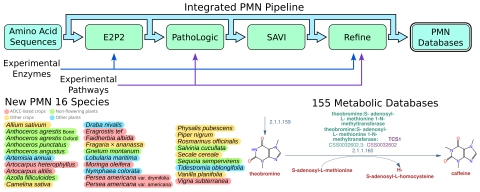
We are super excited to announce the release of PMN 16! PMN 16 introduces twenty-eight new single-species databases, listed below, as well as new versions of the existing 127 single-species databases and to PlantCyc, bringing our total to 155. Selection of new species was focused on understudied plants and plant groups, including non-angiosperm plant such as hornworts and gymnosperms, as well as plants listed by the African Orphan Crop Consortium such as teff, moringa, and bambara groundnut. All databases were (re)generated using our new internal pipeline, making use of Pathway Tools 28.0, the new E2P2 version 5, and updated SAVI validation lists (SAVI 3.2). The first new enzyme data from our collaboration with the labs of Hiroshi Maeda (UW-Madison) and Philipp Zerbe (UC Davis) are also incorporated into this release, with more to come!
E2P2 version 5, the ensemble prediction software developed by the PMN team to predict enzyme function from amino acid sequence, is now available on github (link), and features a significant back-end overhaul. It is now possible to swap out enzymy predictors and to add your own by writing a configuration file. The default predictors are now BLAST and DeepEC, the latter having been swapped in for PRIAM, the second predictor in previous releases. Along with E2P2v5, a new version of the reference protein sequence dataset (RPSD), the set of enzymes collected by the PMN team to use as the basis for E2P2's predictions, has been released. RPSD 5.2 can be downloaded here.
PMN 16 is the first release generated using our new internal pipeline. This update greatly improves the speed, scalability, and reproducability of the pipeline, and now that it is finished we plan to use it to scale up future releases, increasing both size and frequency. In addition, later this year we will, for the first time, be releasing our pipeline to the public. When released, this software will be able to generate pathway genome databases for plant and green algal species using the same procedure that PMN uses. Users will be able to generate their own databases, reproduce our results, and see how the databases change when input options are changed. We're excited to see the new pathway databases people will create!
Finally, we have been collaborating with the labs of Philipp Zerbe and Hiroshi Maeda as part of a new Enzyme Consortium, with the goal of elicidating the function of more members of large classes of enzyme. Our collaborators have already begun work to characterize enzymes in the terpene synthase (TPS), aminotransferase (AT), and cytochrome P450 (CYP) families, and some early results are already incorporated into some of the PMN databases. We look forward to a fruitful continued collaboration!
New databases in this release:
GarlicCyc (Allium sativum), Aagrestis_BonnCyc (Anthoceros agrestis Bonn), Aagrestis_oxfordCyc (Anthoceros agrestis Oxford), ApunctatusCyc (Anthoceros punctatus), AangustusCyc (Anthoceros angustus), SweetWormwoodCyc (Artemisia annua), JackfruitCyc (Artocarpus heterophyllus), BreadfruitCyc (Artocarpus altilis), AfilliculoidesCyc (Azolla filiculoides), CamelinaCyc (Camelina sativa), DrabaCyc (Draba nivalis), TeffCyc (Eragrostis tef), AppleRingAcaciaCyc (Faidherbia albida), StrawberryCyc (Fragaria x ananassa), GamNuiCyc (Gnetum montanum), AlyssumCyc (Lobularia maritima), MoringaCyc (Moringa oleifera), PocketWaterLilyCyc (Nymphaea colorata), MexicanAvocadoCyc (Persea americana drymifolia), HaasAvocadoCyc (Persea americana americana), HuskTomatoCyc (Physalis pubescens), BlackPepperCyc (Piper nigrum), RosemaryCyc (Salvia rosmarinus), WatermossCyc (Salvinia cucullata), RyeCyc (Secale cereale), CoastRedwoodCyc (Sequoia sempervirens), VanillaCyc (Vanilla planifolia), and BambaraBeanCyc (Vigna subterranea)
The PMN BLAST server will be updated in a couple of days with the new databases, and downloads of the new databases will also become available soon
These young scientists had the opportunity to see and touch 3-D models of the molecules that produce each smell, and could sort through each model to identify their favorite scent! Before they left the booth, they were able to choose between different fruit-scented stickers that they could take home with them to remember their experience at Science Night.