In order to assess quality of the data in the PMN databases, evidence codes are associated to the pathways and enzymes.
In the case of computationally derived pathways, color coding of the pathway glyph indicates how well supported individual reactions are.
Evidence Codes
All the databases in PMN use a controlled vocabulary of evidence codes. The three main classes (experimental, author statement and computational) of evidence are graphically represented on database detail pages.
The evidence codes are organized into an ontology that is available from the SRI Pathway Tools site.
An icon representing the type of evidence used to support the pathway is shown in the upper right hand corner of each pathway detail page. There are four kinds of icons:
- A flask (experimental evidence) (example)
- A book (a traceable author statement from the literature) (example)
- A computer (computational evidence) (example)
- A curator (inferred by curator) (example)
As an example, take a look at the pathway detail page for dTDP-L-rhamnose biosynthesis II in AraCyc.
- Go to the upper right hand corner of the pathway detail page. In this pathway there are two evidence icons displayed, a flask and a book. More than one evidence type can be used as support.
- The computer represents evidence taken from computational evidence.
- The flask indicates that evidence for the pathway was also based on experimental data from the literature.
- Point the mouse over one of the icons to see a brief explanation of the evidence code.
- Click on the evidence icon to display a definition of the evidence code and a link to the source of the evidence (e.g. the paper in which an experimental assay is described or where the author statement is found).
- In the window describing the specific support for the evidence code, clicking on the name of the publication takes you to the PubMed abstract or to the journal source of the article
Evidence codes for enzymatic activities
Enzymes catalyzing reactions are also annotated with evidence codes. The same codes and icons are used for enzymes and pathways.
Using the same example pathway as before, GABA degradation, zoom in on the pathway using the 'more detail' button.
- To learn more about 4-amino-butyrate transaminase, click on the name of this enzyme in the pathway diagram to go to the enzyme detail page.
- This page lists all the activities catalyzed by the enzyme. Each of the activity catalyzed has an evidence icon in the upper right corner.
- In this example, the first enzymatic activity (adenosylmethionine-8-amino-7-oxononanoate transaminase activity) has a book icon. Clicking on the icon reveals that this function is supported by a non-traceable author statement, and a link is provided to an abstract from a conference.
- The second enzymatic activity (4-aminobutyrate transaminase activity) has a flask icon. Clicking on the icon reveals that there are two papers demonstrating this activity.
Understanding the pathway evidence glyphs
The pathway evidence glyph on the pathway detail page for computationally-predicted pathways is another source of information about how well supported any pathway is.
Note: Pathway glyphs are NOT present for pathways that have experimental, literature-based, or curator support.
In the pathway evidence glyph:
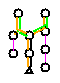
- Each of the reactions in the pathway is color-coded to indicate how much is known about a given pathway.
- Green: An enzyme catalyzing this reaction is present in this organism.
- Black: No enzyme catalyzing this reaction has been identified in this organism.
- Orange: The reaction and any enzyme that catalyzes it (if one has been identified) is unique to this pathway.
- Pink: Represents spontaneous reactions, or lines that do not represent reactions (e.g. in polymerization pathways)
An example of a pathway evidence glyph is seen in the bottom of the Beta-Alanine Biosynthesis I detail page in AraCyc.
- The top two reaction lines are green because enzymes predicted to perform these reactions are present in Arabidopsis.
- The next two reactions in the middle branch of the pathway are black because no enzymes have been identified for these steps in Arabidopsis.
- Orange lines also connect these four reactions, indicating that the reactions shown here are unique to the beta-alanine biosynthesis I pathway. (see note below).
- Three of the reactions along the side branches are pink. Because they are spontanous reactions, no enzyme assignment will be made
The pathway evidence glyph helps to determine whether a computationally-predicted pathway should be retained in the database.
- For example, if an enzyme catalyzes a reaction that is unique to a pathway and is found in the species, this may represent stronger evidence for the existence of that pathway compared to an enzyme that functions in multiple pathways.
The pathway evidence glyph helps to identify so-called "pathway holes" where the enzymes which catalyze specific reactions have not yet been identified.
- Scientists aware of these holes may be able to design experiments to identify the missing components.
Please contact us if you have any questions or check our FAQs page for answers to common questions.



