New CURE Course Offered at UC David Led by PMN Member Philipp Zerbe
The new CURE course at UC Davis, led by PMN member Philipp Zerbe, is helping students to create an interactive chemical map of Arboretum plants. Read about it here: https://biology.ucdavis.edu/news/whats-leaf-students-map-chemistry-campus-plants Photo: Katrina Huynh – UC Davis
New Publication: “Mapping multi-substrate specificity of Arabidopsis aminotransferases”
A new paper by PMN members Hiroshi A. Maeda, Sueng Y. Rhee, Charles Hawkins, and Marcos V. V. de Oliveira, was recently published in Nature Plants. Titled “Mapping multi-substrate specificity of Arabidopsis aminotransferases,” this paper lays the groundwork for engineering crops that use…
PMN Attends Science Night at Glencairn Elementary School
Glencairn Elementary School brought science to their students in East Lansing last week when they held their annual Science Night. This event brings together different members of the science community to showcase to students how science is an integral part of…
The Secret Life of Plant Chemistry: Scents, Flavors, and Survival Tricks!
Join us at the MSU Science Festival on April 5 and 6, 2025, and step into the fascinating world of plant chemistry at our interactive booth! Did you know that tiny molecules called metabolites help plants fight off diseases, attract…
PMN 16 in Focus: New Publication Available Now
We are thrilled to to announce the publication of an article featuring the latest release of the Plant Metabolic Network (PMN 16) in Nucleic Acids Research! This comprehensive update includes over 1,200 metabolic pathways, 1.3 million enzymes, 8,000 metabolites, and…
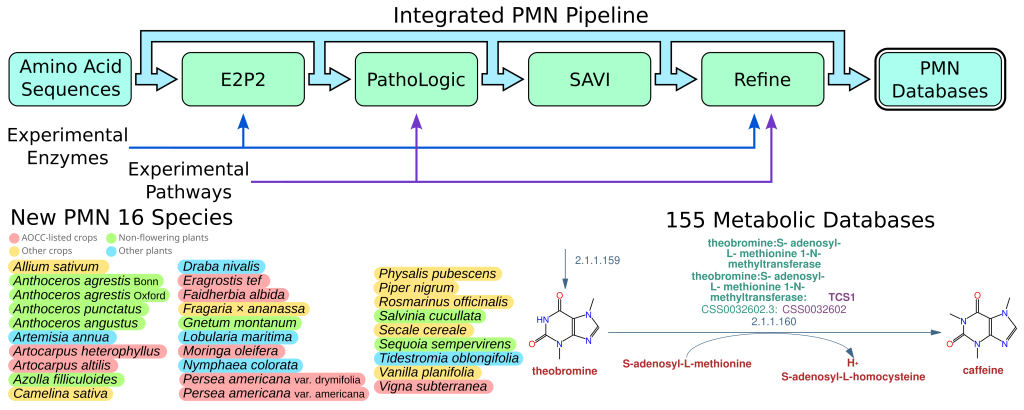
The PMN Pipeline has been released
PMN has been serving the plant science community for more than fifteen years, but until now the resource has been limited to the species databses created by the PMN team using our in-house database createion pipeline. Now, however, for the…
PMN 16 Released!
We are super excited to announce the release of PMN 16! PMN 16 introduces twenty-eight new single-species databases, and new versions of the existing 127 databases, for a total of 155. Selection of new species was focused on understudied plants…
PMN down for updates 7/31
Edit 2: All databases are back up again Edit: The downtime has been pushed to 7/31, same time. PMN will remain up the evening of 7/30 Original post: The PMN databases (meaning all functionality on https://pmn.plantcyc.org) will be down for…
A-Peeling Adventures at the MSU Science Festival
The Plant Metabolic Network (PMN) once again left a lasting impression at the recent STEAM Expo Days event during the MSU Science Festival. Led by Charles Hawkins, director of PMN, a dedicated team of volunteers including Danielle Hoffman, Matt Stata,…
PMN at Glencairn Elementary School Science Night
February 7, 2024 was Science Night at the local Glencairn Elementary School here in East Lansing, MI, and PMN was there with a booth, run by PMN director Dr. Charles Hawkins! We called our booth “Why do bananas smell so…
PMN Receives $3 Million NSF Grant, Big Things Ahead!
Big news! PMN has just received a new grant from the National Science Foundation’s Plant Genome Research Program (PGRP). Worth $3 million over four years, this grant will allow us to significantly expand PMN in size, scope, and usefulness. Big…
PMN 15.5 Released!
We are pleased to announce the release of PMN 15.5! In this release, twenty-five of our databases have been regenerated using updated genome assemblies. We have also updated our backend to Pathway Tools 26.0 and made minor fixes to several…
PMN Downtime March 15
Update 2: The PMN databases are now back up. We thank you for your patience during the move. Update: Due to unexpected difficulties with the server move, PMN downtime has been extended into March 16th. We expect to have PMN…
PMN 15.0 Released!
We are pleased to announce the release of PMN 15. This release includes a new species, new features, and fixes to existing species: We have released a new pathway genome database, StoraCyc, for the legume Senna tora PMN now uses…
Downtime – July 8th
PMN Databases will be offline on July 8th due to datacenter maintenance
New feature: Metabolic Cluster Viewer
We are pleased to announce the launch of a new website feature, the Metabolic Cluster Viewer. We have run our PlantClusterFinder software on nine of the PMN species’ genomes, and the Cluster Viewer allows you to search and view the…
PMN 14.0 released!
We are pleased to announce the PMN14 release. In this release we introduce 25 new organism-specific metabolic pathway databases, which brings the total number of organism pathway databases to 125! New species include ten crops, six wild crop-relatives, three ornamentals,…
PMN13.0 released!
We are pleased to announce the PMN13 release. In this release we introduce 24 new organism-specific metabolic pathway databases, which brings the total number of organism pathway databases to 100! New species include nine crops, two medicinal plants, four ornamental…
PMN12.5 released
We are pleased to announce PMN12.5 release. In this release we added locus IDs to genes in all PMN databases. The locus IDs were extracted from public genome releases that were used in constructing the PMN metabolic pathway databases. Protein…
PMN 12.0 released!
We are pleased to announce the PMN12 release. In this release we introduce 54 new species-specific metabolic pathway databases, which brings the total number of species-specific databases to 76. The 76 species cover the green plant lineage widely, including green…
PMN 11.0 released!
We are pleased to announce the PMN11 release. Highlights include the addition of curated information of rate-limiting steps in over 50 pathways. To see an example, follow this link, http://pmn.plantcyc.org/PLANT/NEW-IMAGE?type=PATHWAY&object=PWY-695 To see a complete list of pathways with rate-limiting info,…
PMN 10.0 released!
We are pleased to announce the PMN10 release. In this release, we introduce 5 new metabolic pathway databases for species including tomato, potato, two diploid progenitors of the bread wheat, Aegilops tauschii (D-genome) and Triticum Urartu (A-genome), and the great…
PlantCyc 9.5 and PapayaCyc 3.5 release
We are pleased to announce PlantCyc 9.5, which fixes a bug in PlantCyc (version 9.0), which was released in September 5, 2014. This bug was caused by internal ID conflict affected only P. patens and C. papaya enzyme annotations in…
PMN 9.0 provides enhanced predictions of enzymes, reactions and pathways!
We improved enzyme and reaction prediction using our new version Ensemble Enzyme Prediction Pipeline (E2P2 v2.1) and update all the 17 species-specific databases. AraCyc (Arabidopsis) BarleyCyc BrachypodiumCyc CassavaCyc (cassava, yucca) ChineseCabbageCyc ChlamyCyc (a green alga) CornCyc GrapeCyc (wine grape) MossCyc…
CornCyc 4.0 adds new content
We identified and fixed a technical data-processing error in CornCyc 3.5.Our updated CornCyc 4.0 contains: 24 new pathways 839 new enzymes 173 new reactions The corrected and updated database, CornCyc 4.0, plus seventeen other databases are available online and can…
PlantCyc 8.0 joins 17 species-specific databases at the PMN: Grasses galore and much more!
Grasses galore and much more! We continue to roll out our PMN 8 release that started in June: New additions: PlantCyc 8.0 (350+ plant species) AraCyc 11.5 (Arabidopsis) ChlamyCyc 3.5 (a green alga) CornCyc 3.5 Released in June 2013:Seven new…
PMN 8.0 provides 17 species: Grasses galore and much more!
Grasses galore and much more! Seven new databases: BarleyCyc BrachypodiumCyc ChineseCabbageCyc OryzaCyc (rice) SetariaCyc SorghumBicolorCyc (sorghum) SwitchgrassCyc Ten updated databases: AraCyc (Arabidopsis) CassavaCyc (cassava, yucca) ChlamyCyc (a green alga) CornCyc GrapeCyc (wine grape) MossCyc (Physcomitrella patens) PapayaCyc PoplarCyc SelaginellaCyc (a…
Get your data groove going with Groups!
Get the data you want in the format you need using the Groups functions at the PMN. * Got a list of up-regulated genes? * Want to know all of the compounds present in the pathways they act in? *…
Finding your favorite gene in AraCyc just got easier!
Now you’re doubly likely to find Arabidopsis genes using AGI locus codes (e.g. AT4G33030) AND Gene symbols (e.g. SQD1) because the PMN has loaded the latest set of gene “symbols” from TAIR into AraCyc. Now over 4600 symbols are linked…
Come see the PMN at PAG in January
The PMN invites you to come to our workshop at the Plant and Animals Genome Conference (PAG XXI) on Sunday, January 13 in San Diego. The PMN, the Sol Genomics Network (SolCyc), and PMN users will present an overview of…
From fruit trees to freshwater algae – FOUR NEW SPECIES at the PMN provide comparative power!
Once again, the Plant Metabolic Network is growing and this time it’s expanding beyond flowering plants with four new databases: ChlamyCyc MossCyc SelaginellaCyc PapayaCyc Come see how our new databases can help you explore metabolism in a broader comparative context.
Desktop downloads offer opportunities
Did you know that you can download all eleven of the PMN databases plus the Pathway Tools software? Working on a local copy of a database enables you to: Build your own pathways View graphs of Omics data directly on…
Plant metabolism grabs the spotlight
Check out this special Science issue on Plant Metabolism Peruse more than 10 reviews, reports, perspectives, and even a podcast to learn about some of the most recent high impact research on medicinal compounds, carbon and nitrogen metabolism, hormonal modifications,…
Come meet the PMN in Austin!
Want to learn about the newest PMN data? Eager for a demo on data analysis using the Omics viewer? Interested in annotating your favorite enzyme? Then meet with a PMN curator one-on-one at the Outreach Booth (215) during the ASPB…
Pondering Molecular Network Evolution
Current Opinion in Plant Biology asks . . . How do the evolutionary forces that act on phenotypes affect the molecular networks that underly plant growth and development? How can researchers elucidate the functions of multi-genic modules? Come read about…
BLAST into four new species!
Deciphering the function of an unknown protein? Searching for a related enzyme in a plant species? Want access to a trusted set of enzymes? Then come BLAST against our new enzyme datasets for corn, soybean, cassava, and wine grape. Plus,…
Podcasting from the PMN!
Rev up your plant metabolism excitement by watching or listening to Episode 78 of This Week in Science and Education where the PMN’s very own Dr. Sue Rhee describes how plant biochemical pathways are the Engine of Life! Do you…
Welcome to FOUR NEW species!
The PMN is pleased to provide four new species-specific databases as part of the PMN 6.0 release: CassavaCyc 1.0 CornCyc 1.0 GrapeCyc 1.0 SoyCyc 2.0 And don’t miss the new data that we have added in AraCyc 9.0, PoplarCyc 4.0,…
Intern input connects you to compounds
We are grateful to our undergraduate interns who have enriched our new and “old” databases with references citing evidence of many compounds found in each species. Check out the bottom of each pathway page to connect to these pertinent publications. …
The PMN goes to Amsterdam!
Please come hear Dr. Sue Rhee, the PI of the PMN project, as she describes genome-wide molecular networks at the Plant Genome Evolution conference in Amsterdam on Tuesday, September 6. And to catch a glimpse of the next generation of…
We want your data at the ASPB Meeting!
Please come to a database overview and hands-on community curation workshop at the ASPB Meeting co-hosted by the PMN. We want your help to get the lastest information on plant metabolism into our resources. PMN/TAIR/SGN Workshop August 7 7:00 PM…
Better Metabolic Over-viewing!
Use the improved Metabolic Overview to easily overlay experimental data find and highlight compounds, enzymes, and reactions zoom and pan against the backdrop of the Arabidopsis or Poplar metabolic map! Whenever you are viewing a single species database like AraCyc…
New PMN releases!
Please come search for your favorite pathways in the newest versions of our PMN databases: PlantCyc 5.0 AraCyc 8.0 PoplarCyc 3.0 With over 40 new pathways and 150 new enzymes, the PMN will help you gain new insights into plant…
Two Gordon Opportunities!
Interested in Plant Metabolic Engineering? There are two great chances to delve into this topic in July 2011! Gordon Research Seminar (July 23-24) *** Abstract submission deadline for oral presentation: March 23 Gordon Research Conference (July 24-29) Please let us…
PMN presentations at a click
Have you attended a PMN workshop or presentation and wanted to review the material? Did you miss a PMN talk at a recent conference and want to catch up? Have you been looking for a quick way to find out…
Come visit the PMN at PAG
There are many ways to connect with a PMN curator at the Plant and Animal Genome XIX Conference in San Diego, California. You can attend a computer demo featuring speakers from three groups who provide plant metabolic pathway databases, read…
New PMN BLAST datasets!
Are you looking for an enzyme in Arabidopsis or poplar that is similar to your favorite protein? Please check out the two new datasets that are available for BLAST analyses: AraCyc Enzymes PoplarCyc Enzymes Our original Reference Enzymes and PlantCyc…
New PMN releases!
Please come search for your favorite pathway using the newest version of the Pathway Tools software in the newest versions of our PMN databases: PlantCyc 4.0 AraCyc 7.0 PoplarCyc 2.0 An introduction to the updated databases was presented at the…
Read all about it . . . in the new PMN paper
Do you want a better understanding of how new PMN databases get predicted from genome sequences? Would you like to read about potential ways to put the PMN to work in your research project? Please come check out our recent…
New viewer for a global metabolic map
New viewer for a global metabolic map Do you want to highlight a compound or reaction on a global metabolic map for a single species? Would you like to overlay your transcript expression, proteomic, or metabolomic data onto a zoomable…
PlantMetabolomics.org and the PMN
The PMN is eager to work with scientists who want to do metabolic-pathway-based analyses.PlantMetabolomics.org is a data repository for a series of metabolomic experiments performed in Arabidopis thalianaYou can read about efforts to incorporate their experimentally identified compounds into AraCyc…
New PMN releases! – PlantCyc 3.0, AraCyc 6.0, and PoplarCyc 1.0
The Plant Metabolic Network is pleased to announce new releases of the metabolic pathway databases AraCyc 6.0 and PlantCyc 3.0, plus the inaugural release of PoplarCyc 1.0 This PMN release includes many updates to AraCyc enzyme/reaction assignments based on a…
Pungent Plant Metabolism – Live!
Please come watch an amazing and enormous Amorphophallus titanum flower bloom . . . and be thankful that you can’t smell it. You can use the PMN to get more information about the stinky metabolites (putrescine and cadaverine) that “Trudy”…
PMN Summer Road Shows!
Many researchers and students from around the world had a chance to see PMN presentations and posters at conferences and universities during the summer of 2009. CIGRAS / UCR July 27-31 San Pedro, Costa Rica American Society of Plant Biologists…
PlantCyc 2.0 and AraCyc 5.0 release!
These releases contain many new pathways, enzymes, reactions, and compounds! A few highlights . . . PlantCyc 2.0 has a whole new set of predicted enzymes from Medicago, tomato, and rice mapped onto the metabolic pathways and more . .…
New easy access to PlantCyc species
Have you ever wondered what information PlantCyc has about apples, bluebells, or carrots? To find out more, please come visit the newly created PlantCyc species page found on the “Databases” submenu. Quickly link to all of the pathways with curated…
New PRIMe Metabolomics Links
Come see new links to metabolite identification resources and data analysis tools from PRIMe on our Metabolomics Resources page. While you’re there, explore the array of helpful websites you can access through these pages.
PMN launches PlantCyc 1.0
On June 17, 2008, PlantCyc made its web debut, containing biochemical pathways from over 250 plant species. The multi-organism database brings together more than 2000 reactions, and includes a total of over 500 pathways.
PMN Roadshow . . . come make connections at upcoming conferences
An important part of the PMN mandate is to encourage interactions and data exchange within the metabolic research community. So, please come meet the PMN staff, give us feedback, and learn about how PMN can help your research at the…
Carnegie Canada Kick-off Jamboree!
Members of the PMN welcomed collaborators from the University of Calgary (David Liscombe, Jeorg Ziegler, Peter Facchini), the Canadian National Research Council – Plant Biotechnology Institute (Dustin Cram, Jonathan Page (via WebX)), and Cornell University (Anuradha Pujar) to an organizational…