Each type of data (Pathway, Reaction, Enzyme, and Compound) in PMN databases has a detail page which can be accessed by the various search functions or through hyperlinks from other data detail pages. This section describes the Pathway Detail, Reaction Detail, Enzyme Detail, and Compound Detail pages.
Each pathway is represented on detail page in a zoomable graphical format along with links to information about the reactions, reactant and product compounds, enzymes and genes.
Pathway Diagram: Each pathway contains some of the following color-coded information.
- Reactions are shown in blue lines, enzymes in yellow, genes in purple, compounds are red.
- Green arrows indicate related pathways (e.g. pathways that feed into or depend upon the currently viewed pathway); click on the arrow to link to the pathway detail page for the related pathway.
- Each of the data types in the pathway diagram are linked to their own detail pages.
Zoom Level Controls: The upper portion of the pathway detail page has two buttons which allow you to zoom in (or out) on the diagram.
- Click on the "More Detail" button to zoom in and show more information about the pathway.
- When the highest level of resolution is displayed, only the "Less Detail" button will be active.
Evidence: Each pathway is associated with an evidence code which indicates the type of data used to support the existence of the pathway. For more specific information, see the tutorial on evidence codes.
- The evidence code icon is shown in the upper right hand corner of the pathway detail page.
- More than one type of evidence may be associated with a pathway. Click on the icon to see details about the evidence and links to the citations where the supporting evidence was derived.
Superclasses: Pathways are classified into a hierarchy of superclasses.
- The superclasses are hyperlinked to a page displaying all of the pathways contained in that superclass. You can navigate up the hierarchy to display similar pathways.
Species Data Available for: It is assumed that the entire pathway presented in the diagram is present in all of the species listed.
- If there are variants on the pathway present in other organisms, these are listed by the Variants heading below the Summary.
Summary: Pathway detail page include a brief summary composed by the database curators.
References: For pathways that have been curated, the literature used as references are hyperlinked in this section.
- Clicking on the citation will display the references used to curate the pathway.
- Click on the reference to see details about the reference.
Unification Links: This section contains links to other databases containing related information.
- The links depend on the type of data being displayed. For example, on a pathway detail page, there may be links to the same pathway in other data bases where you can find curated data about the same pathway in other specieses.
Pathway Evidence Glyph: The pathway evidence glyph is displayed only for pathways that have been computationally predicted.
- The glyph is a symbolic representation of the evidence supporting the individual reactions within a pathway.
- **Even if a pathway diagram shows a flask icon (indicating the pathway is has experimental support), not all reactions in a pathway may be equally well supported.
- The key below the glyph shows the correlation between the color codes and the evidence.
The reaction detail pages can be accessed directly from the search results (e.g. when searching for a specific reaction) and from the list browser. They are also linked from related pathway, compound and enzyme detail pages.
- To access specific reaction details from the pathway detail page, click on the blue reaction line (#426786) corresponding to the pathway of interest.
Superclasses: The reaction hierarchy represented in PMN databases is derived from the Enzyme Commission (EC) classification.
- Each superclass is linked to a list of all of the reactions within the category. Users can browse the hierarchy to locate related reactions.
Pathways: Each reaction detail page lists all of the pathways in which the reaction is known or predicted to occur.
- More than one isozyme can catalyze the reaction; there is currently no correlation between specific isozymes and the pathways in which they act.
Reaction Diagram: A diagram of the reaction is displayed in accordance with the direction of that reaction within a pathway where the substrate compounds are shown to the left of the equation and the product compounds to the right.
- Clicking on the name or chemical structure of any compound will open the compound detail page.
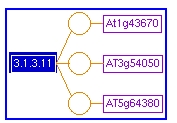
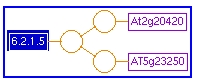
Gene-Reaction Schematic: The gene-reaction schematic is a graphical representation of the relationship among sets of genes, enzymes and reactions.
- The blue box on the left indicates the reaction (e.g. EC 3.1.3.11).
- One or more yellow circles are linked to the reactions. These circles represent polypeptides and protein complexes.
- Purple boxes which represent the genes that encode the proteins are linked to the yellow circles.
- In the gene-reaction schematic provided above, the circles directly connect the genes to the reaction, indicating that the enzyme functions as a monomer.
- In the gene-reaction schematic provided below catalyzed by a protein complex, each gene points to the same circle.
Unification Links: As with the pathway detail page, the unification links point to other databases that describe enzymes, reactions and proteins in the reaction.
The Enzyme detail pages can be accessed directly from the search results, from the list browser and from other detail pages.
- From a pathway detail page, click on the enzyme (yellow text) name(#B07100), or click on the enzyme name on the reaction detail page.
Synonyms: Alternative names for the enzyme are displayed.
- When searching for enzymes by name, the synonyms are also included in the search. Synonyms may be derived from the Enzyme Commissions enzyme nomenclature list.
Superclassess: As with the pathways, reactions and compounds, enzymes are ordered into a hierarchy which can be browsed from this section.
- In general, reactions are classified according to EC hierarchy. Enzymes are not classified; most fall into the polypeptide category.
Comments: Comments include anything that the curators determine are useful information about the enzyme such as specific information about the isozymes, physico-chemical properties (e.g. pH, temperature optimum) and kinetic properties (e.g. Km).
- Clicking on a citation in the Comments section brings you to the list of Citations at bottom of the Enzyme Detail page
- Clicking on a hyperlinked citation in the list of Citations brings you to the PubMed abstract page for the reference, or directly to the journal providing the article.
Species: The species indicates the species(es) in which this enzyme is found.
Gene: The gene(s) that encode for the protein(s) that make up the enzyme are listed.
If an enzyme is multimeric and comprised of different protein products, all of the genes encoding the proteins will be listed here.
Gene-Reaction Schematic: This is the same diagram that is displayed on the reaction detail page.
In the diagram, the enzyme being displayed is indicated by the filled yellow circle.
Enzymatic reaction: The reaction(s) catalyzed by the enzyme are displayed, with the equation set up to reflect the observed directionality.
- It is followed by a list of all the pathways in which this reaction occurs. This information can be useful in determining how changes in a given enzyme may affect metabolic pathways.
- One enzyme may have multiple activities and catalyze more than one reaction. Each activity and the reaction catalyzed are listed on the enzyme detail page along with the evidence supporting the enzymatic activity.
Detailed information about substrate and product compounds can be accessed directly from the database search, or via links from other data detail pages.
Synonyms: Each compound may have one or more synonyms which are alternative names for the compound.
- When searching for compounds in the database, all names including synonyms are included in the search.
Superclasses: Compounds are grouped into classes, which indicate their relationship to other classes of compounds.
- Each superclass is hyperlinked to a list of compounds within that class. This list can be used to find similar compounds to any reactant or product.
Empirical formula and molecular weight: Basic information about the compound such as its empirical formula and molecular weight are shown here. This information can be useful for analyzing metabolomic data.
Structure: The structure of the compound is shown.
SMILES Profile: SMILES is widely used as a general-purpose chemical nomenclature and data exchange format.
- This string can be used to find the same compound in other databases that use the SMILES format.
Unification Links: As with the other detail pages, links to similar or related information about the compound are listed here and hyperlinked to the corresponding database entry.
Participation in reactions
- Reactions in which the compound is a reactant: For each compound, all of the reactions in which the compound is a reactant are listed here. Click on any of the reactions to see the reaction details.
- Reactions in which the compound is a product: If a compound is a product of another reaction, the reaction is listed here and linked to the reaction detail page.
- From these two lists it may be possible to identify other pathways that are likely to be affected by the concentration of a given compound. For example, a mutation that affects an enzyme reaction that generates a specific compound may ultimately affect the functioning of a pathway in which that compound is used as a reactant.
Please contact us if you have any questions or check our FAQs page for answers to common questions.